Convolutional neural networks for image analysis in bioinformatics
Objectives: Gain basic knowledge of convolutional neural networks.
Introduction
Convolutional neural networks (CNNs) are one artificial neural network type specifically designed to process data with a grid-like topology, such as an image.
They are beneficial for image analysis tasks in bioinformatics because they can automatically learn features from the data and use them to classify or distinguish different patterns or objects in images.
Convolutional layers
CNNs consist of multiple layers of interconnected "neurons" inspired by the structure and function of neurons in the biological brain. Each layer processes the input data using a set of learnable weights and biases and passes the output to the next layer.
The layers at the beginning of the network, known as the "convolutional layers," extract features from the input data using a technique called convolution.
In a convolutional layer, the input data passes through a set of filters, which are small matrices of weights that detect specific patterns in the data. For example, we might design a filter to detect edges or corners in an image.
The filters are convolved with the input data by sliding them across the image and performing a dot product at each position.
The convolutions result in a set of feature maps representing the presence or absence of the patterns detected by the filters at different locations in the image.
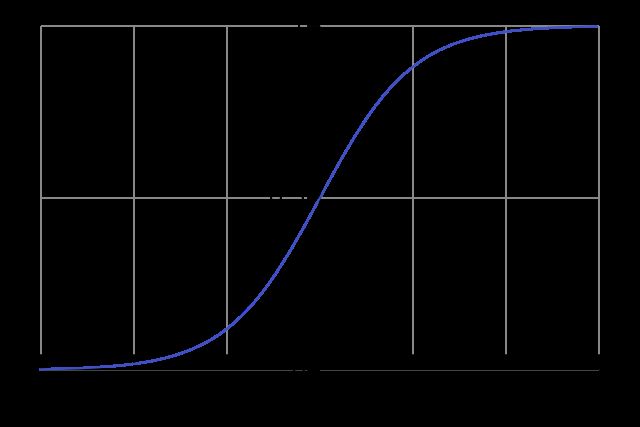
The feature maps are then passed through a series of non-linear activation functions, which introduce non-linearity to the model and allow it to learn more complex patterns. The activation functions are typically applied element-wise to the feature maps, meaning that the activation function transforms each element in the feature map.
Pooling layers
After the convolutional layers, the network typically includes several "pooling layers," which reduce the feature maps' spatial dimensions and increase the model's robustness.
There are several different types of pooling, but the most common is max pooling, which selects the maximum value from a set of adjacent elements in the feature map.
Fully connected layers
Finally, the output of the pooling layers is typically passed through a series of fully connected (or "dense") layers, which use standard neural network techniques to make predictions based on the extracted features.
The output of the final dense layer is the prediction made by CNN.
Training a CNN
In training a CNN, we present the model with a set of training examples, along with the correct labels for each sample. The model uses an optimization algorithm, such as stochastic gradient descent, to adjust the weights and biases of the network to minimize the error between the predicted and actual labels.
This process is repeated for multiple epochs to eventually learn a set of weights that can accurately classify new, unseen examples.
In bioinformatics, CNNs are helpful in various image analysis tasks, including tissue classification, cell counting, and protein localization. They have also been used to analyze medical images, such as MRI and CT scans, for disease diagnosis and segmentation tasks.
Proceed to the next lecture: Autoencoders and their applications in bioinformatics
